Product Description
Also available as a customized product tailored to your needs!
OEM options:
If you need materials as in kit controls or as a third-party control for validation of your kits at customer labs, please contact us via: sales@sens-id.com
How to create your full workflow plasma control material based on this product:
We recommend the use of our human DNA-free plasma products to produce complete workflow controls with this product. Blend the cfDNA as necessary with the DNA depleted plasma to receive plasma materials with the necessary DNA concentrations.

Choose your plasma volume options :
- 5 ml human Plasma (human-tech) 1-fold concentrated
- 40 ml human Plasma (human-tech) 1-fold concentrated
- 1,000 ml human Plasma (human-tech) 1-fold concentrated
These products are ideal for digital PCR and/or Next Generation Sequencing (NGS). In particular:
– Validation and development of sequencing protocols (e.g. Whole Genome Sequencing (WGS), Amplicon Sequencing) and PCR protocols
– Determination of the detection limit of the method
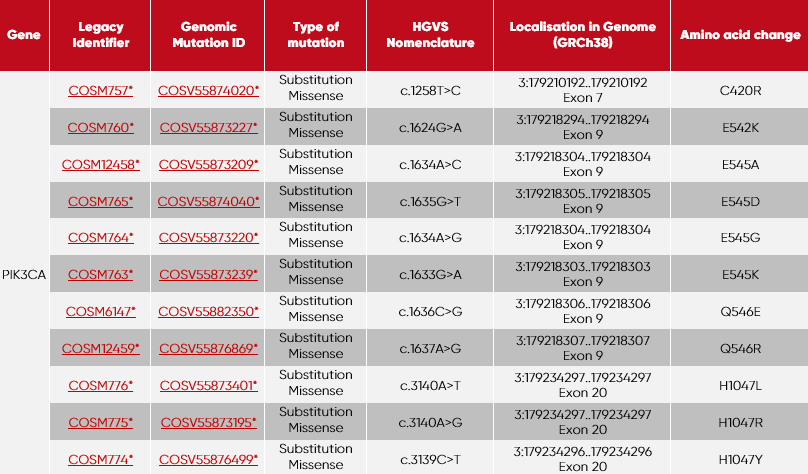
This product contains the following set of tubes:
| Tube | Mutations per tube included |
| 1 | PIK3CA-C420R + PIK3CA-E545A |
| 2 | PIK3CA-Q546R + PIK3CA-E542K |
| 3 | PIK3CA-H1047L + PIK3CA-E545D |
| 4 | PIK3CA-H1047R + PIK3CA-E545G |
| 5 | PIK3CA-H1047Y + PIK3CA-E545K |
| 6 | PIK3CA-Q546E |
| 7 | incl. Wildtype (Ashkenazim son cfDNA Cat. No.: SID-000003) |
Each vial has the following features:
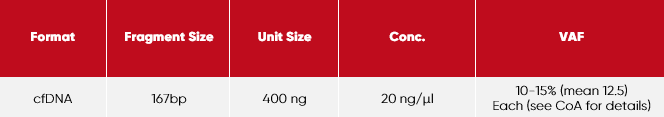
Mutations:
Buffer:
Tris-EDTA (10 mM Tris, 1 mM EDTA), pH 8.0
Storage:
2-8 °C
Expiry:
stable for 24 months from date of manufacturing (as supplied)
Quality control
Fragmentation size:
Electrophoresis-Bioanalyzer-High Sensitivity DNA Kit Agilent
Allele Frequency (metrologically traceable):
ddPCR (Bio-Rad)
Quantification (metrologically traceable):
1. UV-Vis Spectrophotometry (NIST-Reference method)
2. Fluorometric dsDNA measurement (Qubit)
Technical background
Derived from:
The Genome In A Bottle (GIAB) cell line from the Personal Genome Project (PGB): GM24385 (HG002- NA24385 – huAA53E0 – Ashkenazim son)
Bioinformatics:
– Lot specific sequencing files: LOT Search
– High-confidence variant calls: ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio
– Raw datasets and bam files, currently including 10X Genomics, BioNano, Complete Genomics regular and LFR, 300x Illumina paired-end, Illumina 6kb mate-pair, 1000x Ion exome, custom moleculo libraries, ~0.05x Oxford Nanopore, and 70x/30x/30x PacBio: ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/
* GRCh38 · COSMIC v90
Literature:
1. The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61-70.
2. Tolaney S, Toi M, Neven P, et al. Presented at: 2019 American Association for Cancer Research (AACR) Annual Meeting; March 29 – April 3, 2019; Atlanta, GA.
3. Di Leo A, Johnston S, Seok Lee K, et al. Lancet Oncol. 2018;19(1):87-100.
4. Moynahan ME, Chen D, He W, et al. Br J Cancer. 2017;116(6):726-730.
5. Mutation distributions and clinical correlations of PIK3CA gene mutations in breast cancer








